answer 4:1 - least squares fit of Y = [X]/(K+[X])
In theory the answer is correct, and this is the strategy applied in tutorial #1: the use of least squares minimization is the soundest statistical way of analyzing the experimental data. You use the following equation to calculate the fraction of liganded sites:
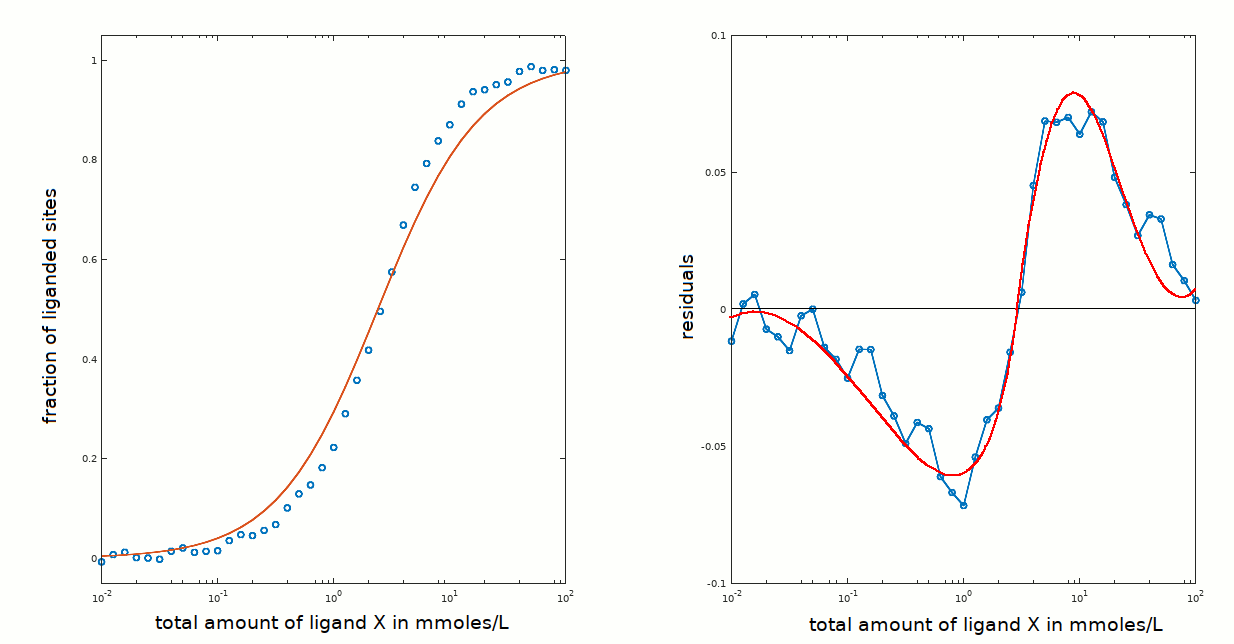
The equilibrium constant obtained is Kd = 2.4 mM (for the dissociation reaction: PX <==> P + X).
The residuals are clearly unsatisfactory, because they show systematic deviations from the zero line (the description of "good" and "bad" residuals' plots has been given in the preceding tutorials). However in this case we know that the protein is monomeric and chemically pure: we cannot invoke positive or negative cooperativity or chemical heterogeneity to explain the above result.
The student is invited to consider the following question before proceeding to the next page: which is the effect of the protein concentrationin this experiment?
Go to the following page
Back one step