answer 3:4 - Analysis of the residuals
The calculated ligand binding isotherm is broader than the experimentally determined one and the residuals do not look good, because they are non-random and cross the zero line at very low frequency, suggesting that the fit has a systematic error.

You can compare the results obtained in this tutorial with those obtained in tutorial #1, where the residuals presented high frequency and no evidence of systematic deviations from the zero line:
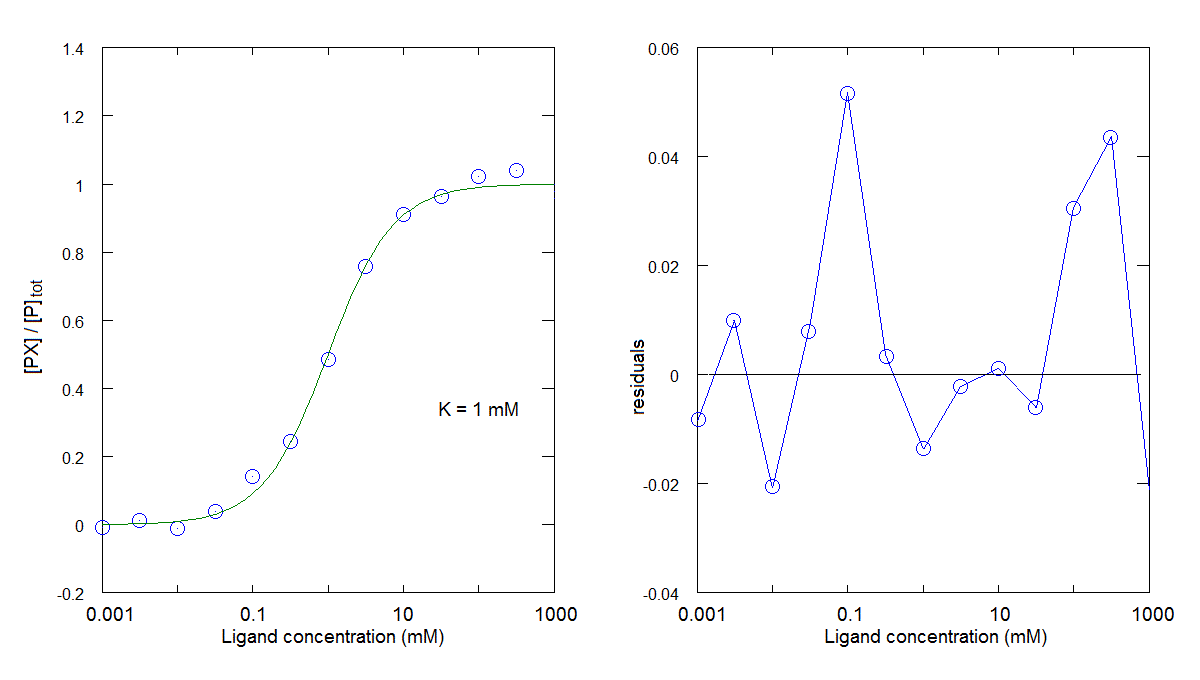
You can also compare the results obtained in this tutorial with those obtained in tutorial #2, where the residuals presented a systematic deviation from the zero line in a direction opposite to the one observed above:
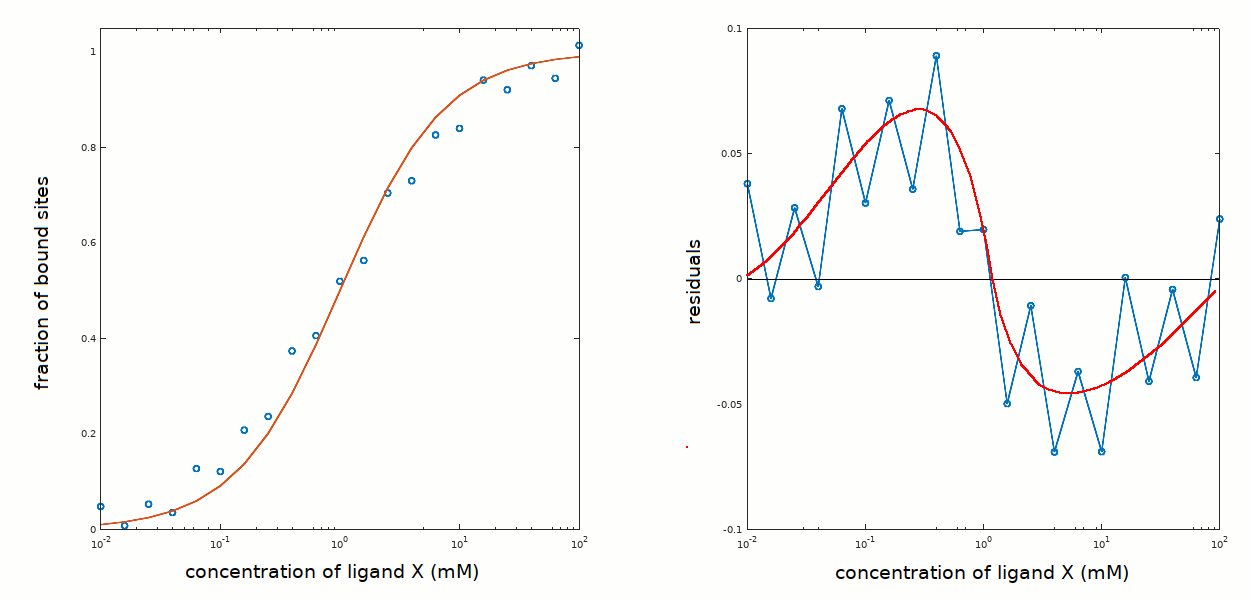
The analysis is unsatisfactory: some new hypothesis should be formulated and tested:
1) The protein is not a monomer and presents positive cooperativity.
2) The protein is not a monomer and presents negative cooperativity.
3) The protein is a mixture of different monomeric isoforms.