answer 1:1 - least squares fit of the signal
Congratulations! This is the statistically soundest procedure to follow.
Unfortunately it is not the easiest to implement. What you have to do is:
1) write down an equation that calculates the signal change as a function of ligand concentration; this must necessarily pass through the ligand binding function but is not limited to it. You use the following equation to calculate the fraction of liganded sites:
2) Use a computer equipped with a non-linear least-squares minimization routine to find the parameters of the above equations (Kd, offset, and Δ signal) that yield the best approximation of the experimental data.
You obtain the following results:
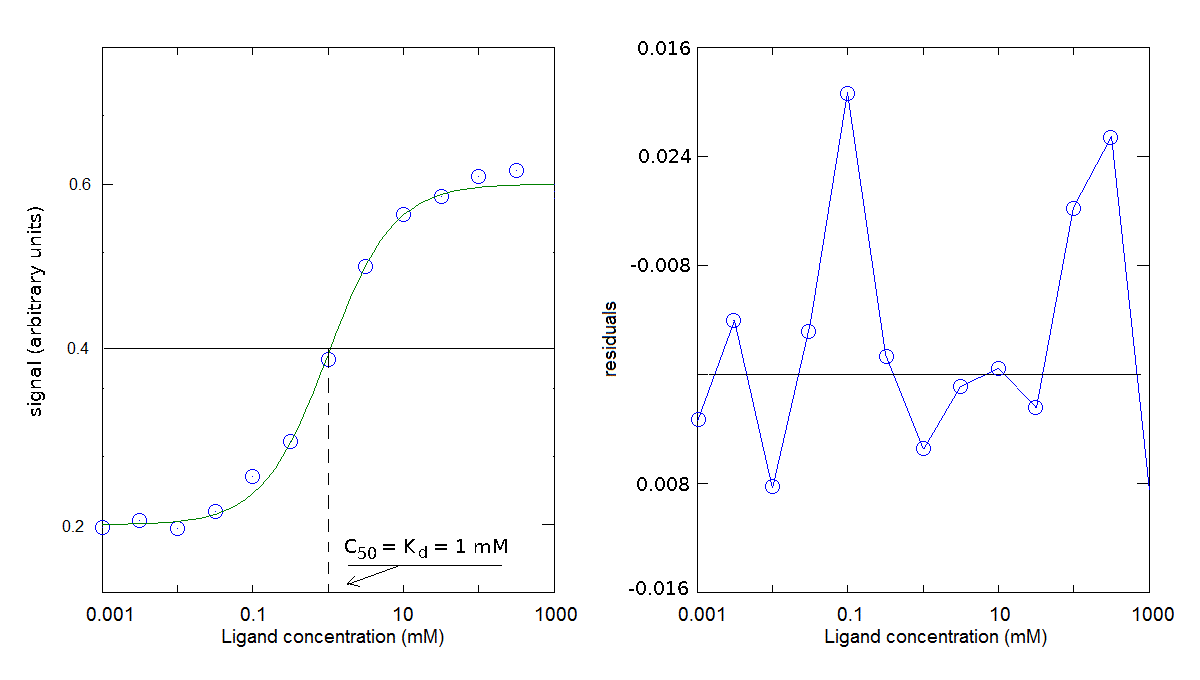
The parameters obtained is Kd = 1 mM (for the dissociation reaction: PX <==> P + X); offset=0.2; Δ signal=0.4.
I remark that the midpoint of the signal change (i.e. the condition [PX] = 1/2 [P]tot) is reached when [X] = Kd. This ligand concentration is called the C50.
Do the residuals look good?
1) Yes
2) No
Back one step